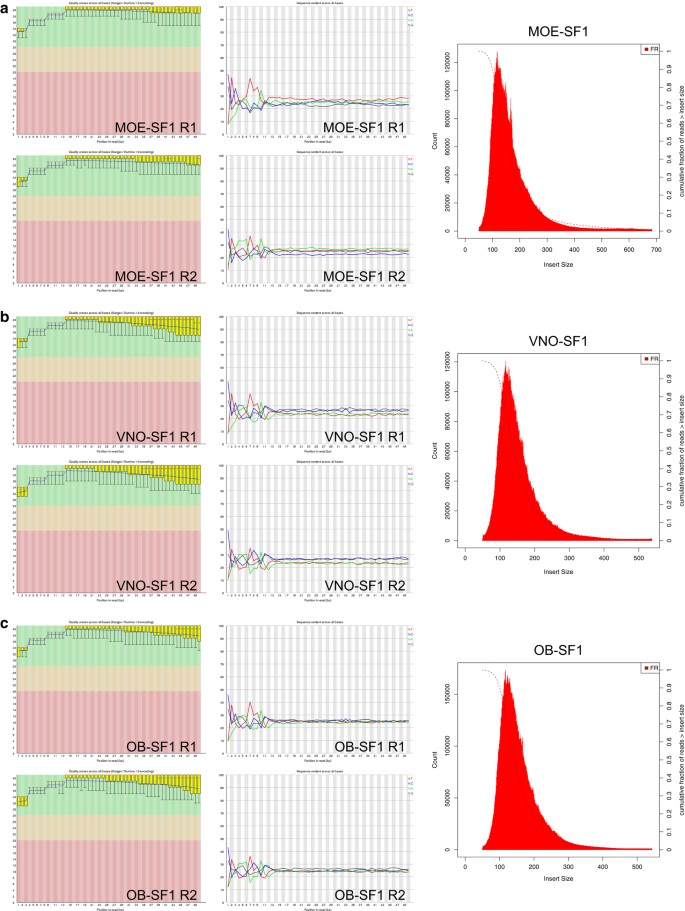
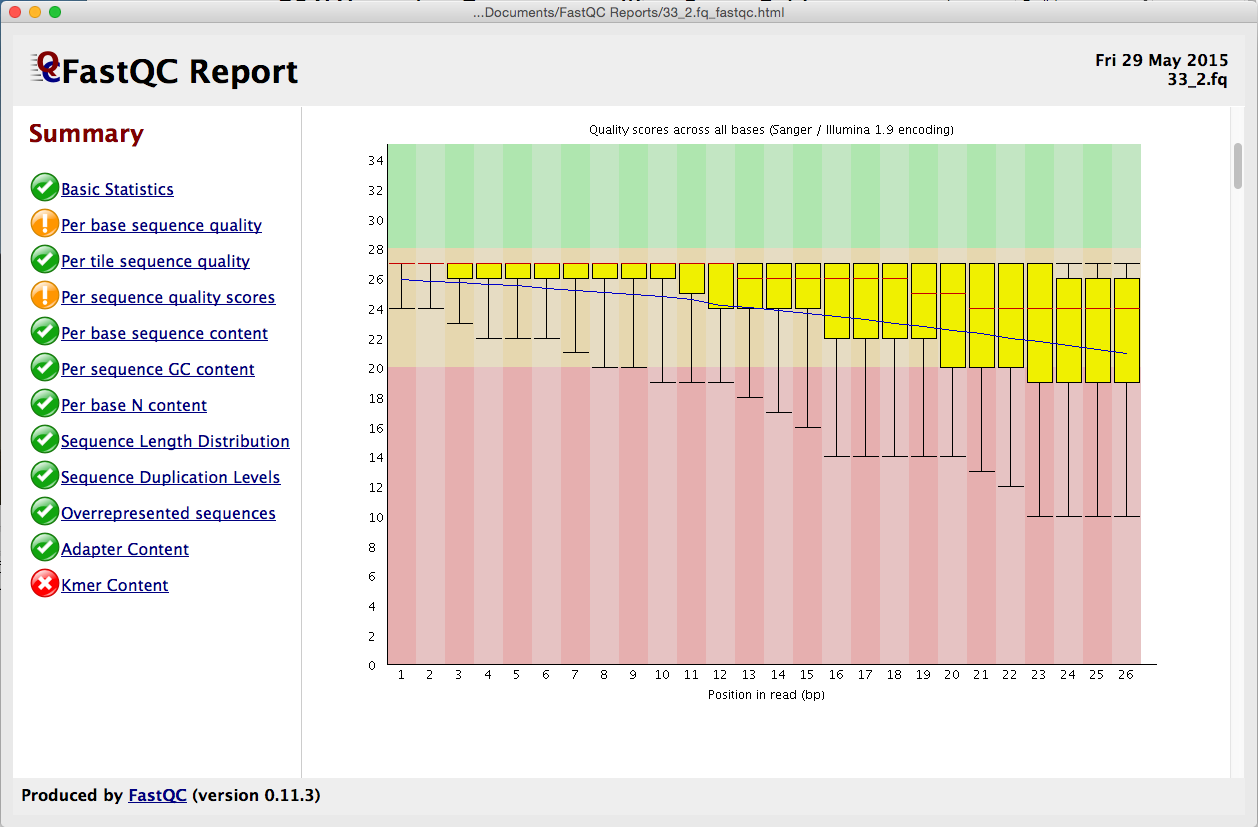
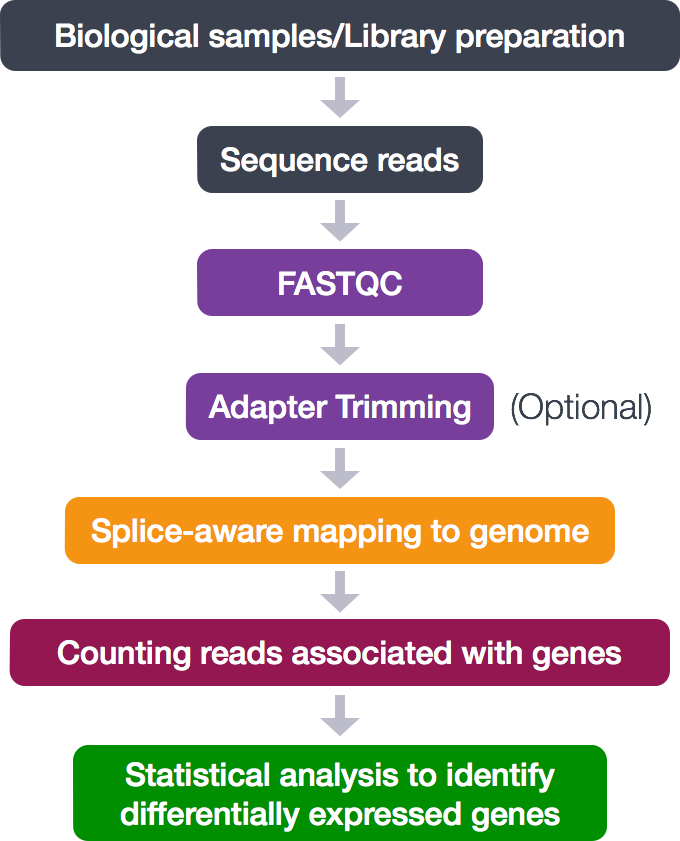
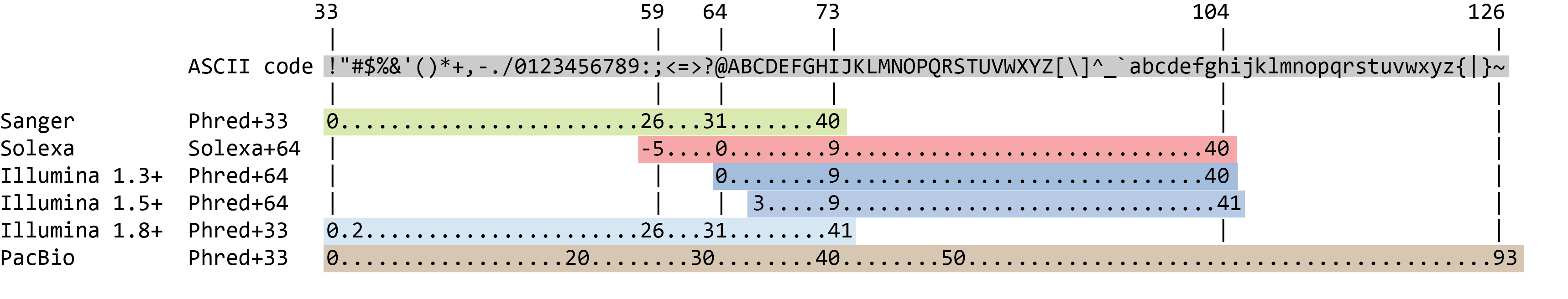
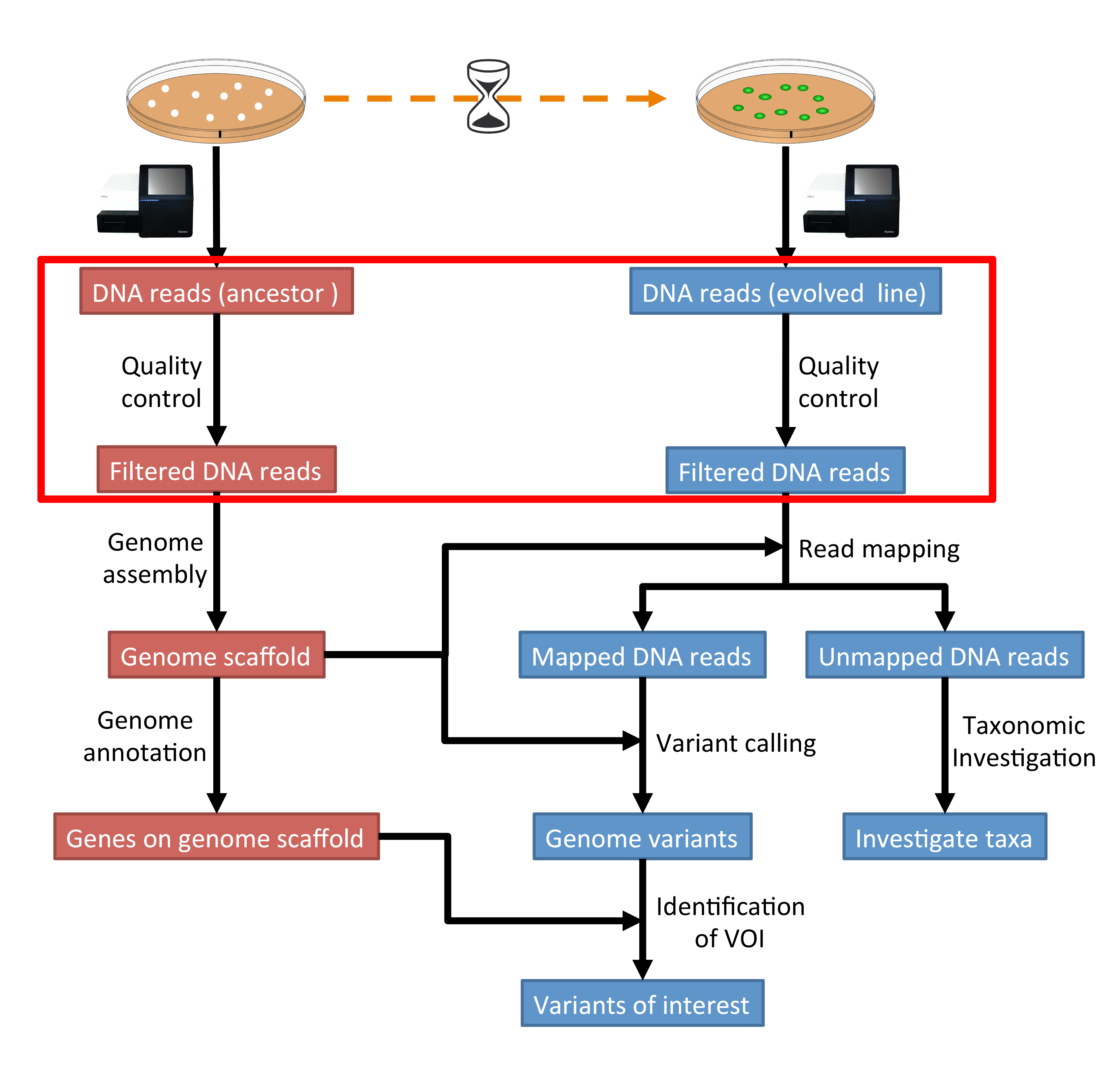
Statistical guidelines for quality control of next-generation sequencing techniques | Life Science Alliance
GitHub - natir/fastQt: FastQC port to Qt5: A quality control tool for high throughput sequence data.
![PDF] FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool | Semantic Scholar PDF] FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/eacf7e5551a8cea85278c2a746714f20258664ea/2-Figure1-1.png)
PDF] FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool | Semantic Scholar